raul-padron.org
"CURIOSITY-DRIVEN RESEARCH RULES"

Superb photograp "The Ants Dreams!" by Rakesh Rocky @rakeshrocky1705
"CURIOSITY-DRIVEN RESEARCH RULES"

Superb photograp "The Ants Dreams!" by Rakesh Rocky @rakeshrocky1705
 |

|
Raúl Padrón ORCID ID 0000-0002-1412-2450  |
|
 |
 |
MRC
LMB Alumnus HHMI Alumnus/activities
TWAS elected member ACAL
elected member NAS
elected international member Biophysical
Society emeritus member PUBLICATIONS
Dr. Raúl Padrón, PhD in Physiology and
Biophysics of IVIC (1979), is Professor at the Department of Radiology,
Division of
Cell Biology and Imaging, University
of Massachusetts Medical School
in Worcester, Massachusetts, U.S.A. and Senior
Emeritus Investigator at the
Center of Structural Biology (CBE) of the Venezuelan
Institute for
Scientific Research (IVIC)
in Caracas, Venezuela, where he started
his
scientifc career as visiting student in 1966. After his postdoctoral
work with Hugh E. Huxley
at the MRC Laboratory of
Molecular Biology (Cambridge, UK) (supported
by the Alberto Vollmer Foundation)
he founded the Deparment
(now Center) of Structural Biology of IVIC
in 1997, where he was an International Research Scholar of the Howard
Hughes Medical Institute (HHMI) from 1997 until 2011. He has
devoted his career
to the study of the structure and function of the myosin thick
filaments of skeletal, cardiac and smooth muscle and the myosin
interacting-head motif (IHM) structure and function, with their
implications on the muscle relaxed state, super-relaxation, thick
filament activation and human muscle diseases like hypertrophic and
dilated cardiomyopathy. His honors includes: Polar
Prize
("Lorenzo Mendoza Fleury" Prize)
(1991) of Fundación
Empresas Polar (FEP); CONICIT
Biology Prizes (1989, 1990, 1996); FONACIT Biology Prize (2005); and
the National Prize in Science and Technology of Venezuela (2008). He is
an elected member of the Latin-American
Academy of Sciences (ACAL) (2002), and The
World Academy of Sciences (TWAS) (2004), and an elected
international member of the National
Academy of Sciences of the U.S.A (2018). | Till November 12,
2018: Center of
Structural Biology (CBE)
Venezuela
Institute for Scientific Research (IVIC) |
|
From November 13,
2018: Padrón
Profile Craig
Lab Radiology
Department University of Massachusetts
Medical School Cryo-EM
Core Facility
|
| |
| Padrón, R., Ma, W., Duno-Miranda, S., Koubassova, N., Lee, K. H., Pinto, A., Alamo, L., Bolaños, P., Tsaturyan, A., Irving, T. and Craig, R. (2020) The myosin interacting-heads motif present in live tarantula muscle explains tetanic and post-tetanic phosphorylation mechanisms. Proceedings of the National Academy of Sciences |
| Toepfer, C.,
Garfinkel, A., Venturini, G., Wakimoto, H., Repetti, G., Alamo, L.,
Sharma, A., Agarwal, R., Ewoldt, J., Cloonan, P., Letendre, J., Lun,
M., Olivotto, I., Colan, S., Ashley, E., Jacoby, D., Michels, M.,
Redwood, C., Watkins, H., Day, S., Staples, J., Padrón, R.,
Chopra, A., Ho, C., Chen, C., Pereira, A., Seidman, J., Seidman, C.
E. (2020) Myosin sequestration regulates sarcomere function,
cardiomyocyte energetics, and metabolism, informing the pathogenesis of
hypertrophic cardiomyopathy. Circulation
2020; 141:828–842. DOI: 10.1161/CIRCULATIONAHA.119.042339. |
| Sulbaran, G,
Biasutto, A., Mendez, F., Pinto, A., Alamo, A. and Padrón, R.
(2020) 18O labeling on Ser45 but not on Ser35 supports the cooperative
phosphorylation mechanism on tarantula thick filament activation.
(2020) Biochem
Biophys Res Comm. 524: 198-204. |
|
L. Alamo, A. Pinto, G. Sulbarán, J. Mavárez & R. Padrón “Lessons from a tarantula: New insights into myosin interacting-heads motif evolution and its implications on disease” Biophys. Rev https://doi.org/10.1007/s12551-017-0292-4. |
| Mendoza,
F. & Padrón, R. La Revolución de la
Resolución: la Crio-Microscopía Electrónica
de partículas aisladas resuelve la estructura atómica de
biomoléculas en
solución. Avances en Química: 13 (1) 7-13, 2018.
PDF |
| Lee K H,
Sulbarán G, Yang S, Mun J Y, Alamo
L,
Pinto A, Sato O, Ikebe M, Liu X, Korn E D, Sarsoza F, Bernstein S I,
Padrón R,
Craig R (2018) Interacting-heads
motif has been conserved as a mechanism of myosin II inhibition since
before
the origin of animals. Proc Natl Acad Sci USA 115:E1991-E2000 https://doi.org/10.1073/pnas.1715247115
PDF
SI
PDF |
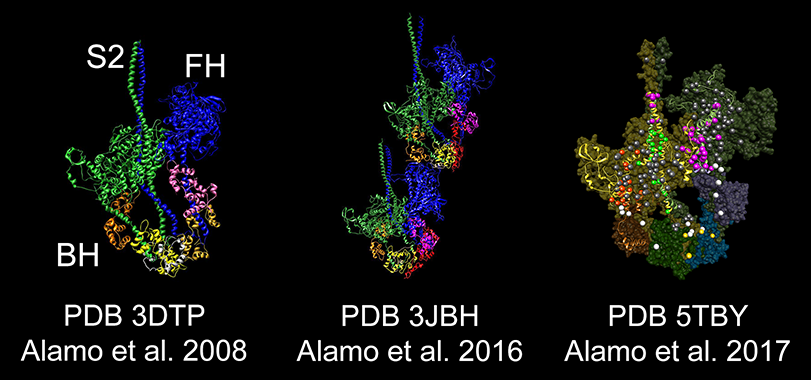
| L. Alamo, N. Koubassova, A. Pinto, R. Gillilan, A. Tsaturyan & R. Padrón 2017 Lessons from a tarantula: New insights into muscle thick filament and myosin interacting-heads motif structure and function Biophys. Rev. 9:461-480 DOI 10.1007/s2551-017-0295-4 SharEdit PDF: http://rdcu.be/vz7T |
| L. Alamo, J. S. Ware, A. Pinto, R. E. Gillilan, J. G. Seidman, C. E. Seidman & Raúl Padrón 2017 Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes eLife 2017;6:e24634 doi: 10.7554/eLife.24634 PDF SI PDF |
| Myosin
II thick filament structure, function, evolution and disease |
Myosin
II interacting-heads motif (IHM) structure, function, evolution and
disease |
| Whis is the structure of relaxed thick filaments? | Which is the structure of the switched OFF IHM? |
| How
the relaxed thik filament structure is conserved? |
How
the relaxed IHM structure is conserved? |
| How thick filaments are relaxed and activated? | How
the IHM is relaxed and activated? |
| How
thick filaments malfunction on disease? |
How IHM malfunction on disease? |

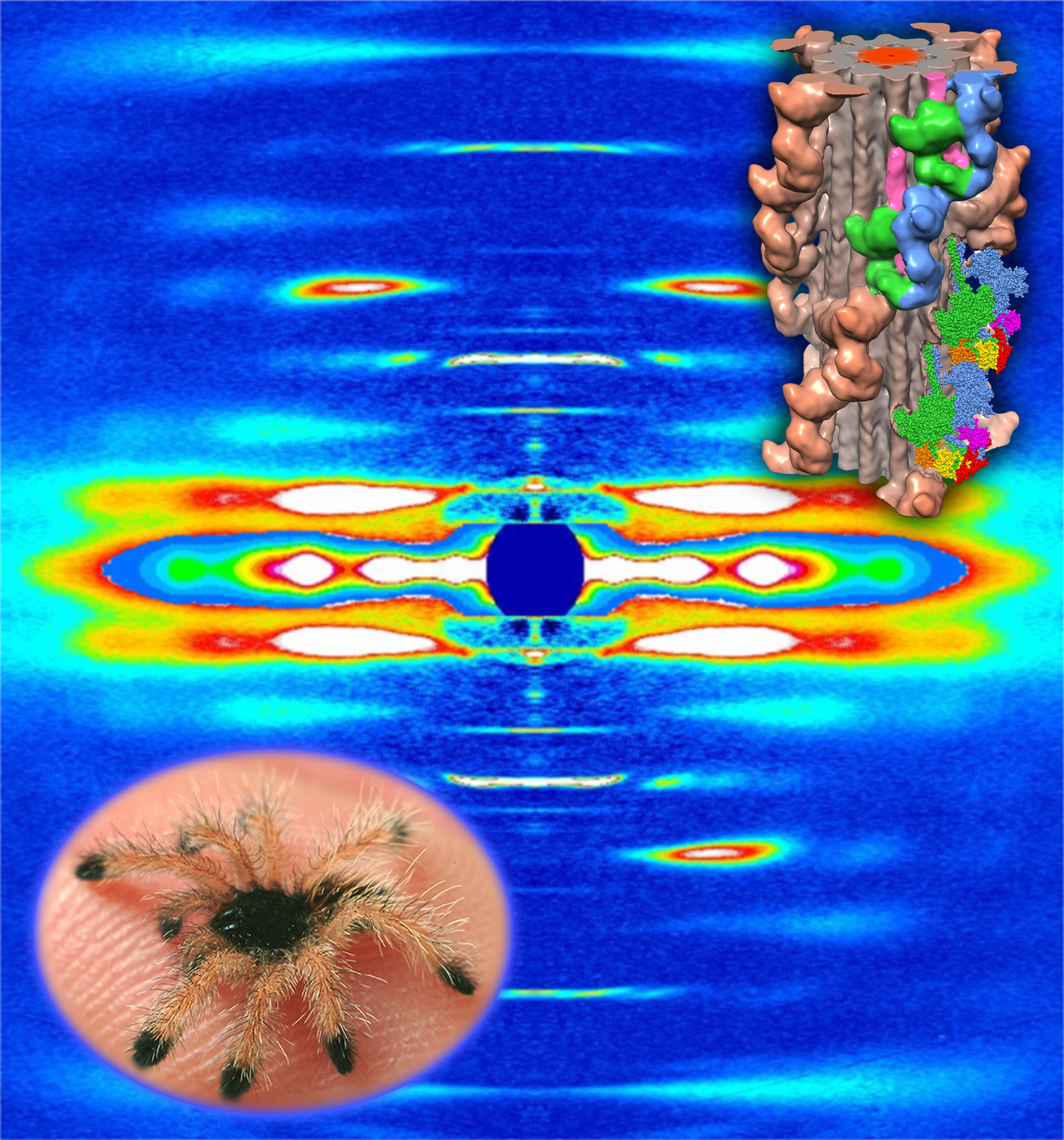
2 nm resolution three-dimensional reconstruction of tarantula striated muscle thick filament
(Solid model 3D-printed by Prof. Dr. Ulrich Meisner, Johannes Gutenberg Universitat, Institute fur Zoologie, Mainz, Germany)
|
Which is the structure
of relaxed tarantula thick filaments
|
||||
|
Structural evidences of
the tarantula thick filament
x
|
||||
| The search for the tarantula thick filament
structure: from 5.0 to 1.3 nm resolution (1985-2016) |
||||
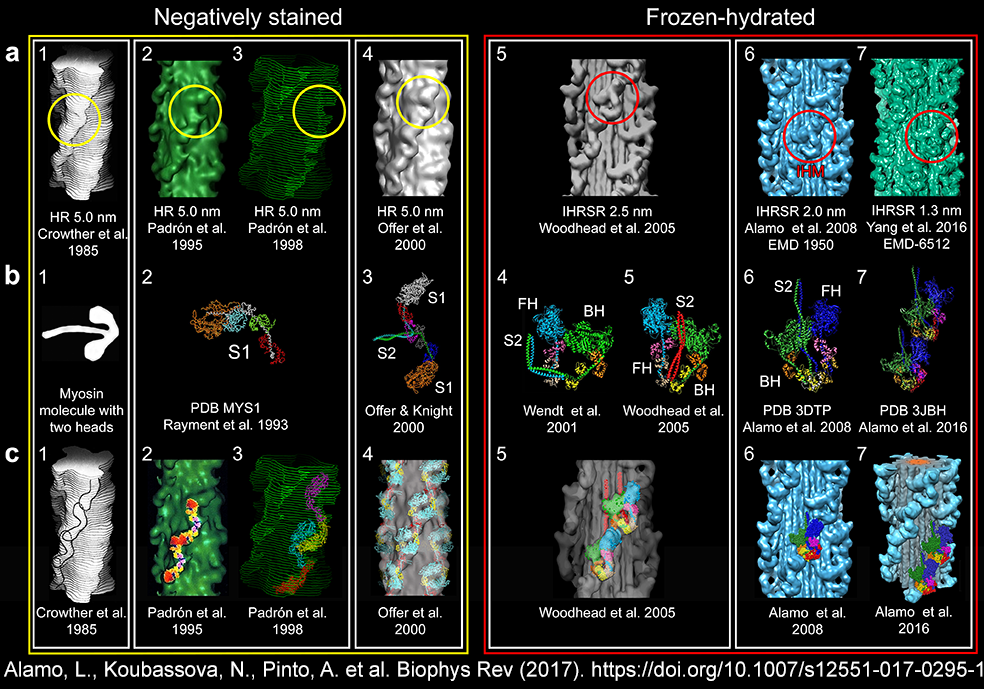 |
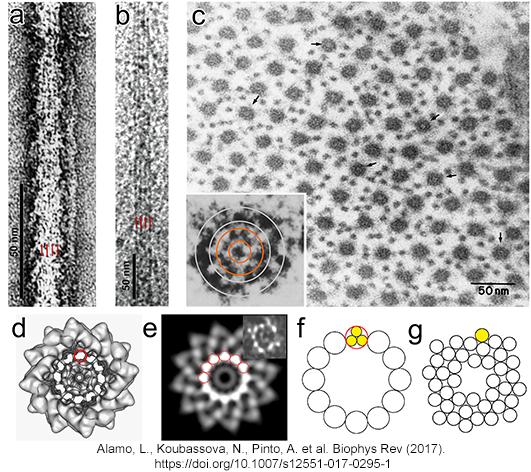
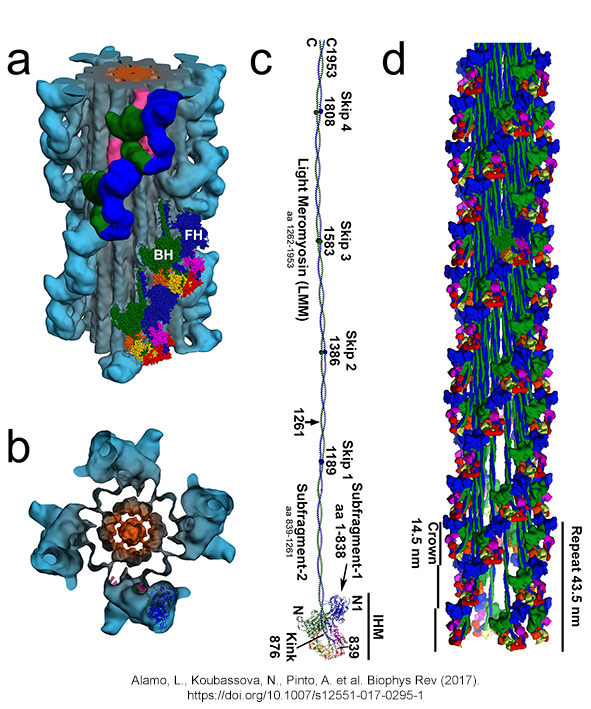
| (a,b)
Tarantula thick filament 3D-reconstruction, (a) IHM quasi-atomic model
PDB 3JBH, (c) myosin II quasi-atomic model and (d) thick filament
quasi-atomic model |
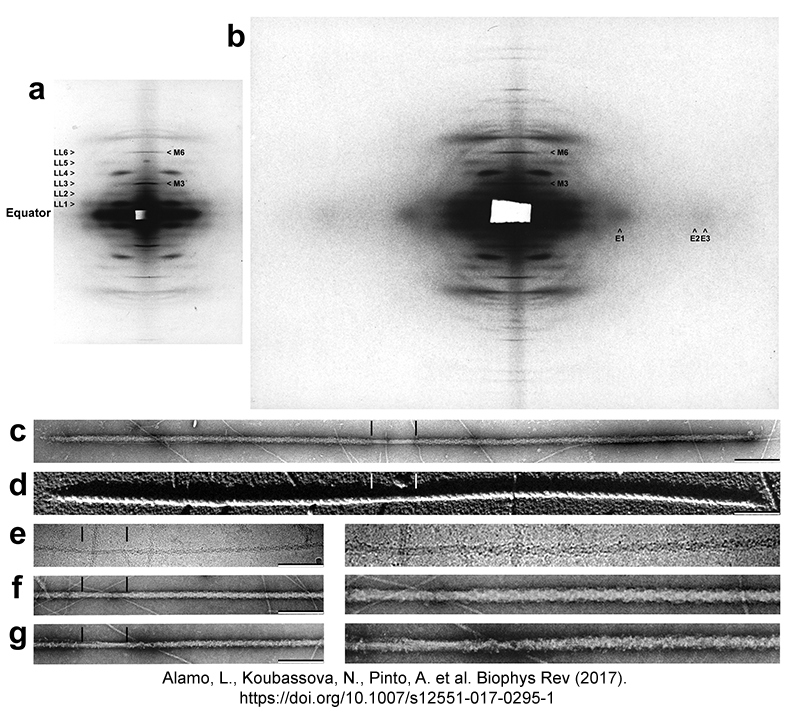
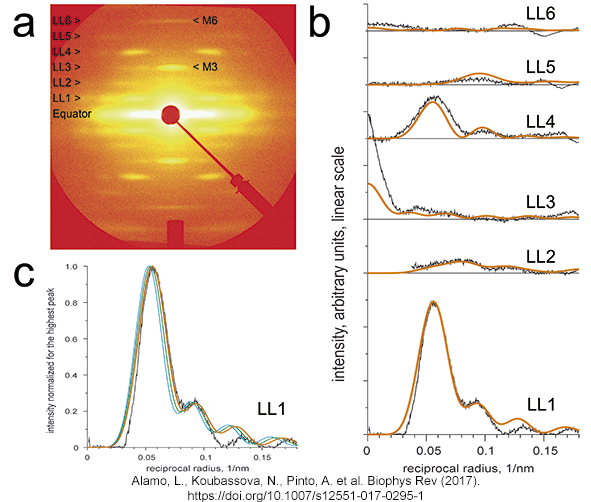
Tarantula
low-angle X-ray diffraction pattern (a) vs. diffraction of the
tarantula quasi-atromic model (b,c) |
 |
 |
|
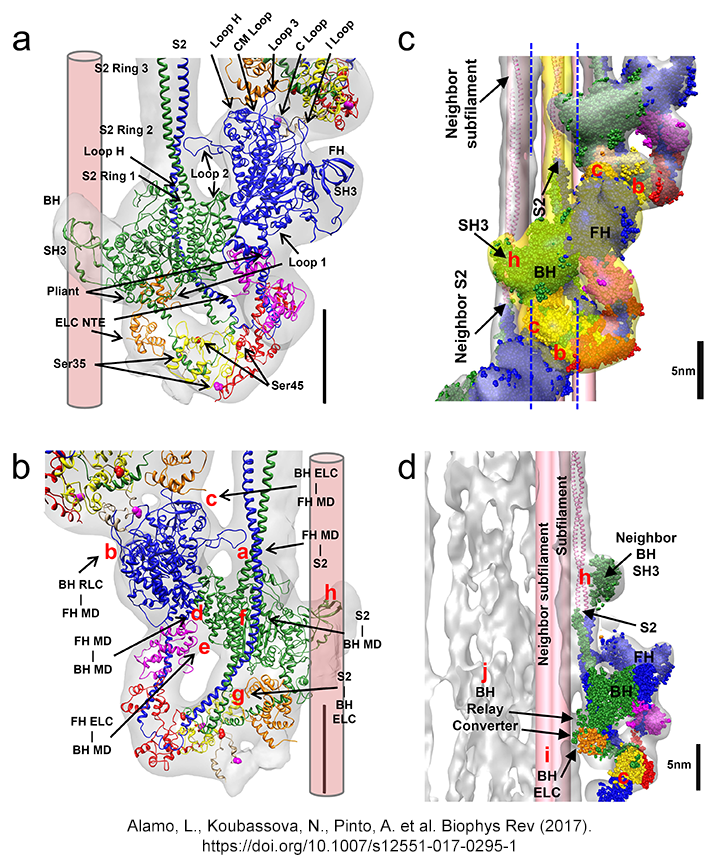
IHM´s Intramolecular and
intermolecular interactions
(Alamo et al. J. Mol. Biol. 2016) DOI 10.1007/s2551-017-0295-4 http://rdcu.be/vz7T |
 |
 |
|||||||||||||||||||||
|
Tarantula myosin II and paramyosin sequences
| Myosin II heavy chain (MHC) |
Myosin regulatory light chain (RLC) | Myosin
essential light chain (ELC) |
Paramyosin
heavy chain (PM) |
||
| Aphonopelma GenBank: KT619079.1 Alamo et al. J. Mol. Biol. 2016 |
|
Aphonopelma GenBank: KT390185.1 Zhu et al. 2009 |
Aphonopelma GenBank: KT692662.1 Alamo et al. J. Mol. Biol. 2016 |
| Swaying heads, potentiation and
postetanic potentiation mechanisms Alamo et al. J. Mol. Biol. 2008, Brito et al. J. Mol. Biol. 2011 |
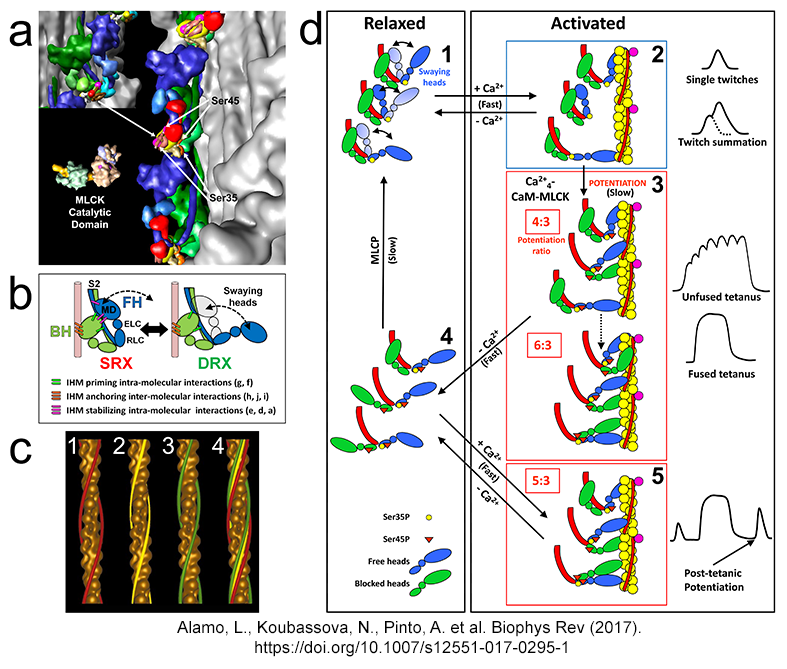 |
| The cooperative phosphorylation
activation (CPA) mechanism of thick filament activation Sulbarán et al. Biophys. J. 2011, Espinoza-Fonseca et al. Mol. BioSyst. 2015, Alamo et al. Mol. BioSyst. 2015, Alamo et al J Mol Biol 2016, Alamo et al. Biophys. Rev. 2017, Sulbaran et al BBRC 2020, Padron et al. PNAS accepted |
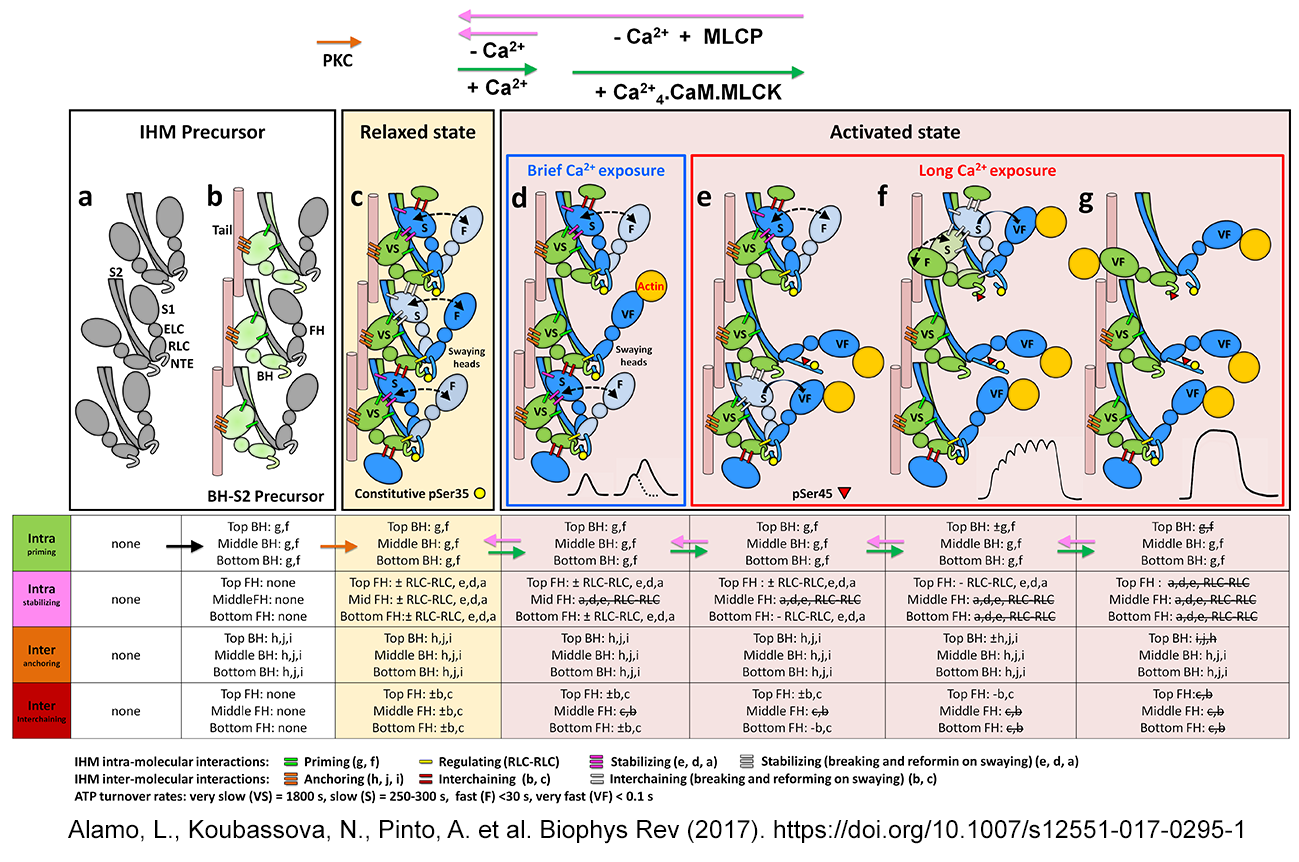 |
| The swaying-swinging, tilting
crosbridge-sliding filament mechanism Alamo et al. Biophys. Rev.2017 |
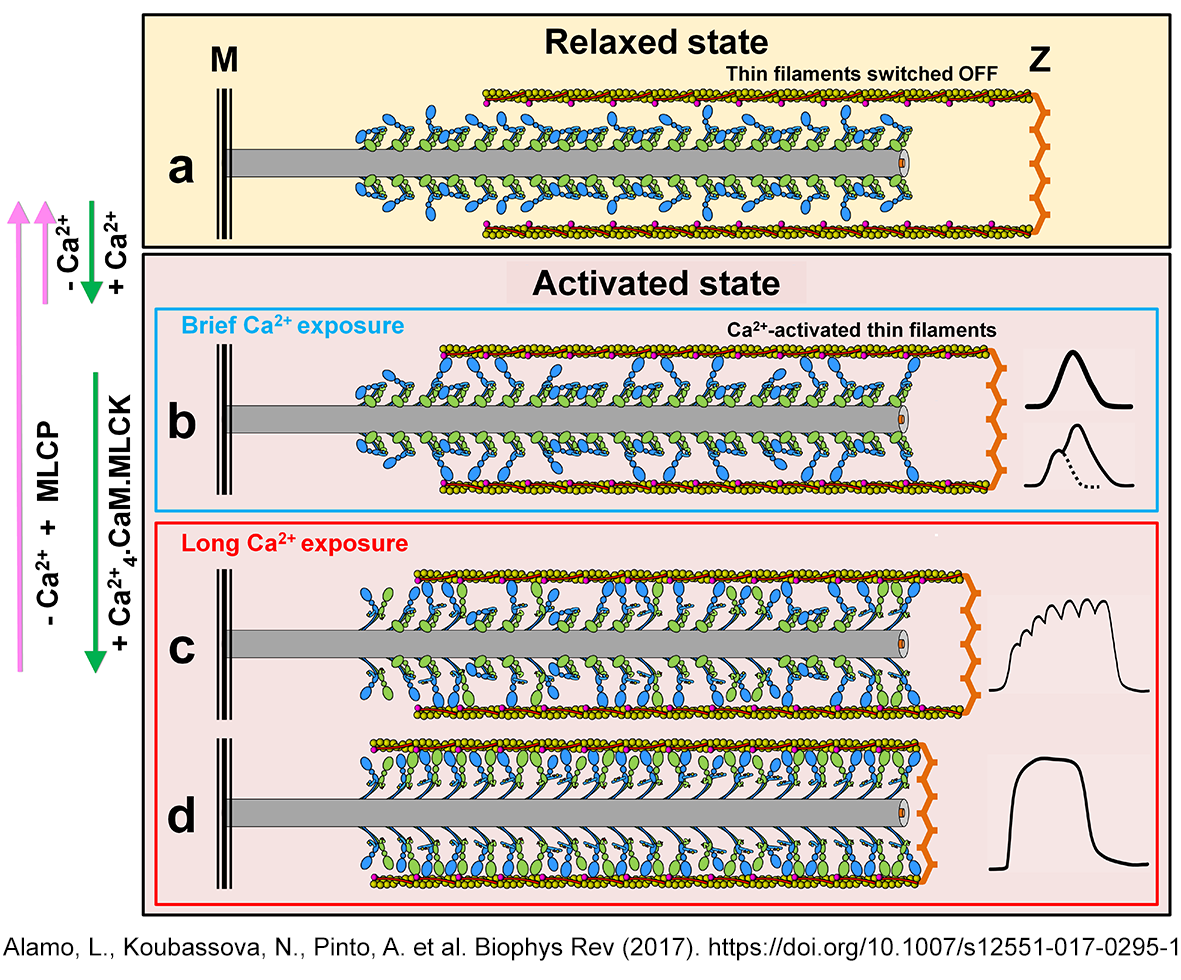 |
Lorenzo Alamo, James S. Ware, Antonio Pinto, Richard E. Gillilan, Jonathan G. Seidman, Christine E. Seidman & Raúl Padrón eLife 2017;6:e24634 doi: 10.7554/eLife.24634
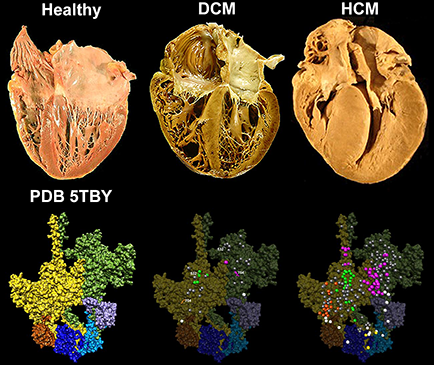 |
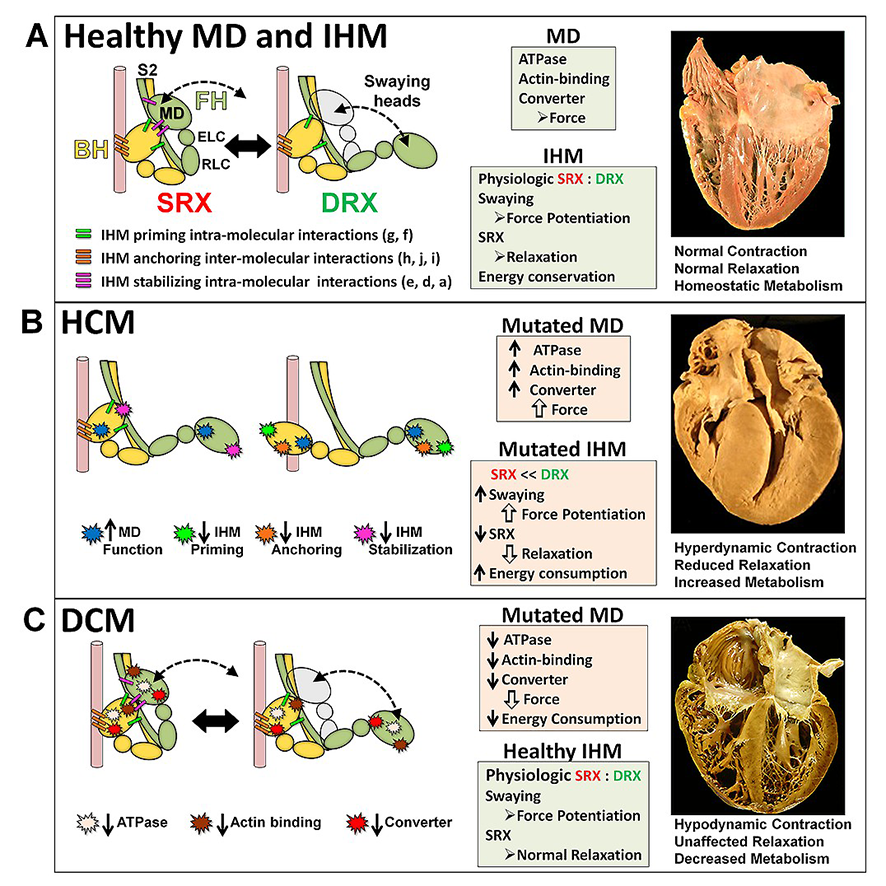 |
Cardiac β-myosin
variants
cause hypertrophic
cardiomyopathy (HCM) or dilated
cardiomyopathy (DCM) by disrupting sarcomere contraction and
relaxation. The locations of variants on isolated myosin head
structures predict
contractility effects but not the prominent relaxation and energetic
deficits that characterize HCM. During relaxation, pairs of myosins
form interacting-heads
motif (IHM) structures that with other sarcomere proteins establish
an energy-saving, super-relaxed (SRX) state. Using a human
β-cardiac myosin IHM quasi-atomic model (PDB
5TBY), we defined interactions sites between adjacent myosin heads
and associated protein partners, and then analyzed rare variants from
6112 HCM and 1315 DCM patients and 33,370 ExAC controls. HCM variants,
72% that changed electrostatic charges, disproportionately altered IHM
interaction residues (expected 23%; HCM 54%, p=2.6×10−19;
DCM 26%, p=0.66; controls 20%, p=0.23). HCM variant locations predict impaired
IHM formation and stability, and attenuation of the SRX state -
accounting for altered contractrility, reduced
diastolic relaxation, and increased energy consumption, that fully
characterize HCM pathogenesis.
|
Howard
Hughes Medical Institute (HHMI) News Lessons
from a Tarantula: Spider muscles reveals details about mutations that
disrupt heart relaxation by Hanae Armitage)
Harvard Medical School News
Lessons
from a Tarantula: Spider muscles reveals details about mutations that
disrupt heart relaxation by Hanae Armitage
Imperial College
London News Spider
proteins offer new insight into human heart conditions by
Ryan O´Hare
MRC
London
Institute of Medical Sciences Biomedical
picture of the day "Spinning spider proteins" by Deborah
Oakley
Christine E. Seidman´s lecture "Hearts, Spiders, and Relaxation: Voyages in Hypertrophic Cardiomyopathy" NHLBI 70th Anniversay - 4-25-2018 Micro radial RCR 750 : "Raúl Padrón y sus tarántulas" de Micros radiales ACFIMAN |
| 1988 |
1991 |
1997 |
1998 |
1995 |
2003 |
2003 |
2008 |
2011 |
 |
 |
 |
 |
 |
 |
 |
 |
 |